Improving 3D CT Organ Segmentation
An AI for Medical Imaging project that improved 3D segmentation of thoracic organs by systematically addressing severe class imbalance and model noise.
More About the Project
Python | PyTorch | Data Analysis | Loss Functions | Jupyter
This project's goal was to improve upon a baseline ENet model for 3D segmentation of organs (Heart, Aorta, Esophagus, Trachea) in CT scans. The primary challenges were severe class imbalance and noisy predictions. I analyzed and implemented various techniques, a key component of this was a deep dive into custom loss functions and data analysis to mitigate imbalance.
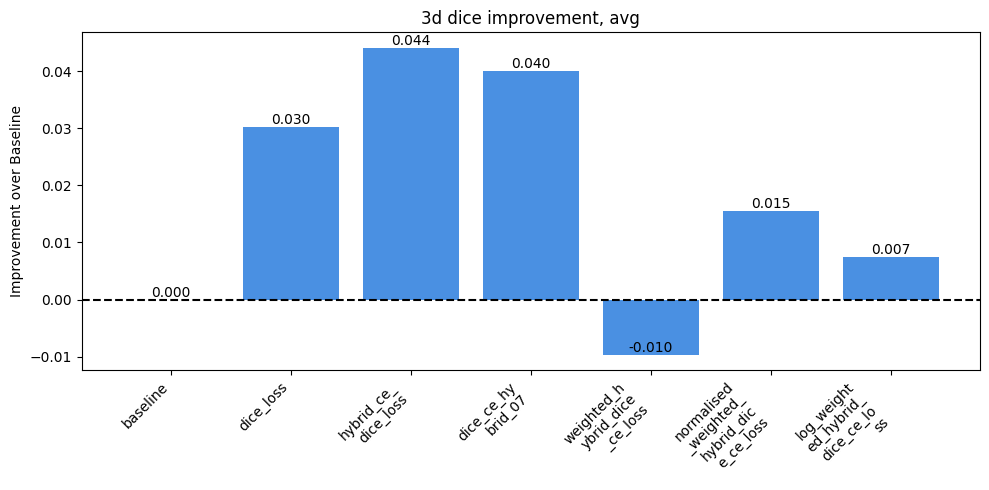
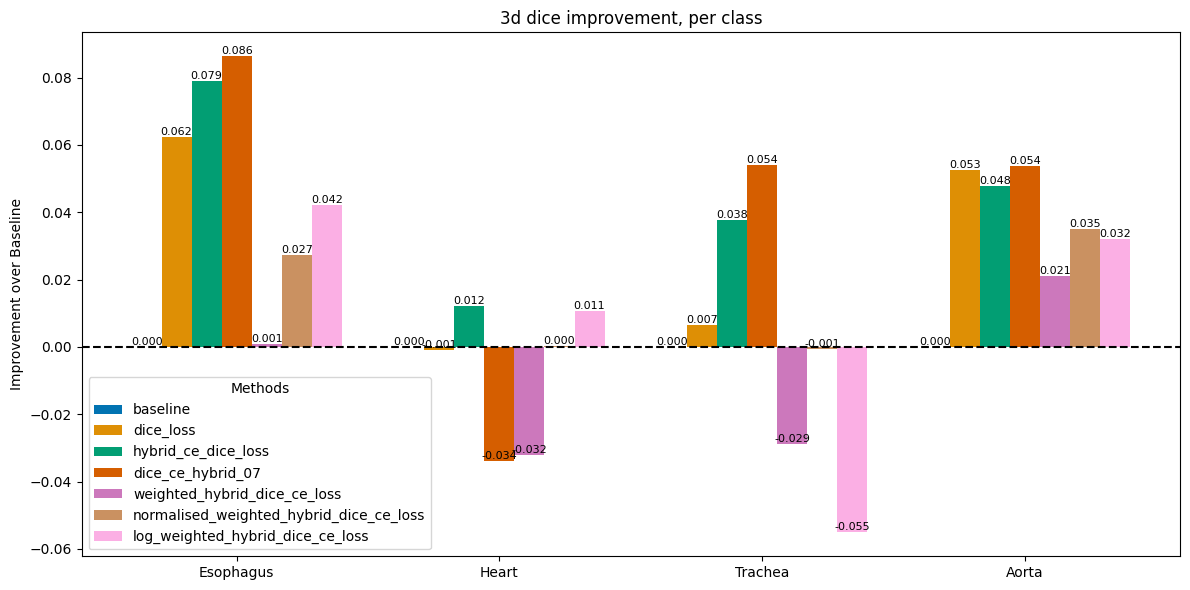
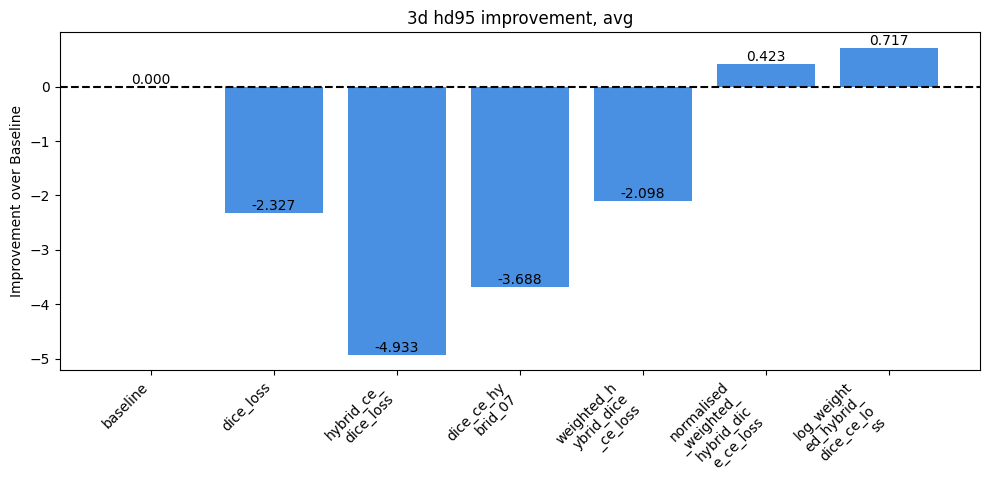
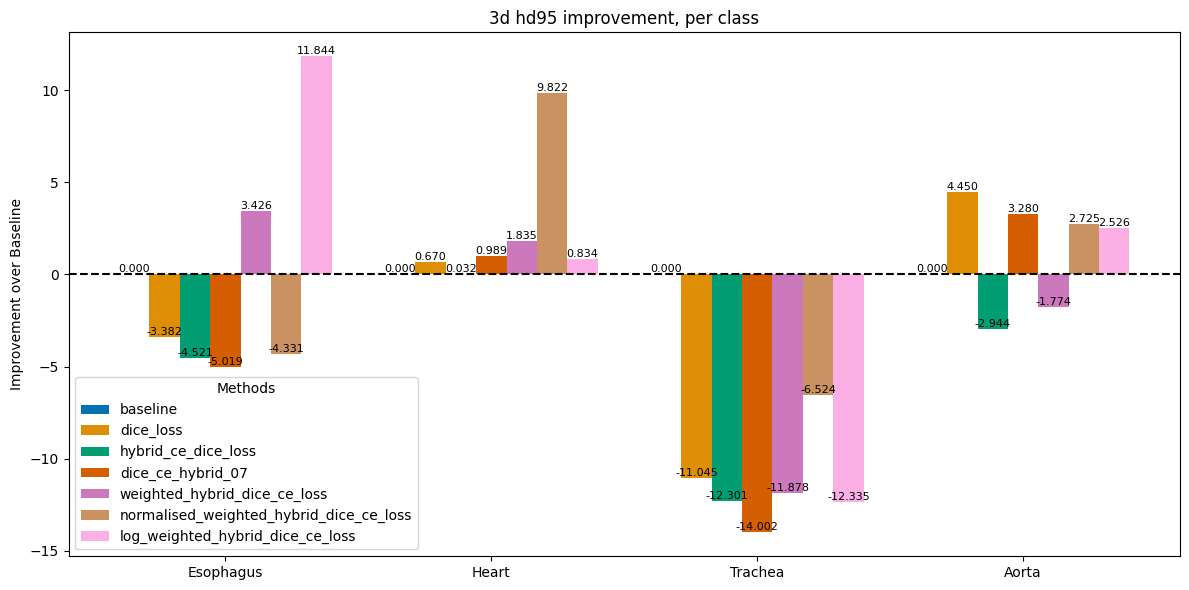
Impact of Loss Functions on Dice & HD95
My Contributions
Data Analysis and Problem Identification
Analyzed the voxel and slice distributions for all 40 participants, identifying severe class imbalance as a core limitation.
Quantified that background voxels made up 98.95% of the data, with rare classes like Esophagus (0.05%) and Trachea (0.03%) being vastly underrepresented.
Loss Function Experimentation
Identified that the baseline cross-entropy loss struggled with rare classes.
Researched and implemented alternative loss functions to address the imbalance, including pure Dice loss and various hybrid (Dice + Cross Entropy) losses.
Analyzed the trade-offs of these losses, noting that Dice loss improved performance on rare classes by focusing on overall shape rather than individual voxels.
Model Improvement and Evaluation
Contributed to the development of the final ‘Best Model,’ which integrated multiple improvements (2.5D, Multi-Window, SE blocks, NAdam).
The final model achieved major gains in rare class segmentation.